Poly-ubiquitination, the process in which a chain of at least four ubiquitin peptides are attached to a lysine on a substrate protein, most commonly results in the degradation of the substrate protein via the proteasome. Apparently, at least four ubiquitins are required on a substrate protein in order for the proteasome to bind and therefore degrade the substrate (though there are examples of non-ubiquitinated proteins being targeted to the proteasome).
Mono-ubiquitination, the process in which a single ubiquitin peptide is bound to a substrate, initiates cell signaling by allowing other proteins that contain ubiquitin binding domains to interact with the mono-ubiquitinated substrate. Mono-ubiquitination has been associated with targeting of membrane proteins to the lysosome, for example.
The ubiquitylation system was initially characterised as an ATP-dependent proteolytic system present in cellular extracts. A heat-stable polypeptide present in these extracts, ATP-dependent proteolysis factor 1 (APF-1), was found to become covalently attached to the model protein substrate lysozyme in an ATP and Mg2+-dependent process. Multiple APF-1 molecules were linked to a single substrate molecule by an isopeptide linkage and conjugates were found to be rapidly degraded with the release of free APF-1. Soon after APF-1-protein conjugation was characterised, APF-1 was identified as ubiquitin. The carboxyl group of the C-terminal glycine residue of ubiquitin (Gly76) was identified as the moiety conjugated to substrate lysine residues.
Contents |
The protein
|
Ubiquitin is a small protein that occurs in all eukaryotic cells. Its main function is to mark other proteins for destruction, known as proteolysis. Several ubiquitin molecules attach to the condemned protein (polyubiquitination), and it then moves to a proteasome, a barrel-shaped structure where the proteolysis occurs. Ubiquitin can also mark transmembrane proteins (for example, receptors) for removal from membranes and fulfill several signalling roles within the cell.
Ubiquitin consists of 76 amino acids and has a molecular mass of about 8500 Da. It is highly conserved among eukaryotic species: Human and yeast ubiquitin share 96 % sequence identity. The human ubiquitin sequence is:
MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG

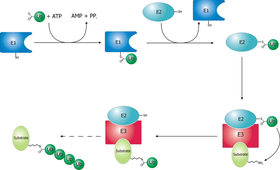
Ubiquitylation
The process of marking a protein with ubiquitin (ubiquitylation or ubiquitination) consists of a series of steps:
- Activation of ubiquitin - Ubiquitin is activated in a two-step reaction by an E1 ubiquitin-activating enzyme in a process requiring ATP as an energy source. The initial step involves production of an ubiquitin-adenylate intermediate. The second step transfers ubiquitin to the E1 active site cysteine residue, with release of AMP. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group.
- Transfer of ubiquitin from E1 to the active site cysteine of an ubiquitin-conjugating enzyme E2 via a trans(thio)esterification reaction. Mammalian genomes contain 20-30 UBCs.
- The final step of the ubiquitylation cascade
generally requires the activity of one of the hundreds
of E3 ubiquitin-protein ligases (often termed simply
ubiquitin ligase). E3 enzymes function as the substrate
recognition modules of the system and are capable of
interaction with both E2 and substrate. E3 enzymes
possess one of two domains:
- The HECT (Homologous to the E6-AP Carboxyl Terminus) domain
- The RING domain (or the closely related U-box domain)
- Transfer can occur in two ways:
-
- Directly from E2, catalysed by RING domain E3s.
- Via an E3 enzyme, catalysed by HECT domain E3s. In this case, a covalent E3-ubiquitin intermediate is formed before transfer of ubiquitin to the substrate protein.
-
In many cases, ubiquitin molecules are further added on to previously-conjugated ubiquitin molecules to form a polyubiquitin chain. If the chain is longer than 3 ubiquitin molecules, the tagged protein is rapidly degraded by the 26S-proteasome into small peptides (usually 3-24 amino acid residues in length). Ubiquitin moieties are cleaved off the protein by deubiquitinating enzymes and are recycled for further use.
Cell-surface transmembrane molecules that are tagged with ubiquitin are often mono-ubiquitinated, and this modification alters the subcellular localization of the protein, often targeting the protein for destruction in lysosomes.
The ubiquitin pathway is thought to be the method of cellular egress for a number of retroviruses, including HIV and Ebola, but the exact mechanism by which this occurs has yet to be deduced.
The Anaphase-promoting complex (APC) and the SCF complex (for Skp1-Cullin-F-box protein complex) are two examples of multi-subunit E3s involved in recognition and ubiquitination of specific target proteins for degradation by the proteasome.
Disease association
Genetic disorders
- The gene whose disruption causes Angelman syndrome, UBE3A, encodes an ubiquitin ligase (E3) enzyme termed E6-AP.
- The gene disrupted in Von Hippel-Lindau syndrome encodes an ubiquitin E3 ligase termed the VHL tumor suppressor or VHL gene.
- The gene disrupted in Liddle's Syndrome results in disregulation of an epithelial Na+ channel (ENaC) and causes hypertension.
Immunohistochemistry
Antibodies to ubiquitin are used in histology to identify abnormal accumulations of protein inside cells that are markers of disease. These accumulations are called inclusion bodies. Examples of such abnormal inclusions in cells are
- Neurofibrillary tangles in Alzheimer's disease
Lewy body in Parkinson's disease
Pick bodies in Pick's disease
Inclusions in motor neuron disease
Mallory's Hyalin in alcoholic liver disease
Rosenthal fibres in astrocytes
External links
Further reading
- Essays in Biochemistry, Volume 41 (2005): The Ubiquitin-Proteasome System (Portland Press)




 216.73.216.133
216.73.216.133 User Stats:
User Stats:
 Today: 0
Today: 0 Yesterday: 0
Yesterday: 0 This Month: 0
This Month: 0 This Year: 0
This Year: 0 Total Users: 117
Total Users: 117 New Members:
New Members:
 216.73.xxx.xxx
216.73.xxx.xxx
 Server Time:
Server Time: