DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light can cause DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day.[1] Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. Consequently, the DNA repair process must be constantly active so it can respond rapidly to any damage in the DNA structure.
The rate of DNA repair is dependent on many factors, including the cell type, the age of the cell, and the extracellular environment. A cell that has accumulated a large amount of DNA damage, or one that no longer effectively repairs damage incurred by its DNA, can enter one of three possible states:
- an irreversible state of dormancy, known as senescence
- cell suicide, also known as apoptosis or programmed cell death
- unregulated cell division, which can lead to the formation of a tumor that is cancerous
The DNA repair ability of a cell is vital to the integrity of its genome and thus to its normal functioning and that of the organism. Many genes that were initially shown to influence lifespan have turned out to be involved in DNA damage repair and protection.[2] Failure to correct molecular lesions in cells that form gametes can introduce mutations into the genomes of the offspring and thus influence the rate of evolution.
Contents |
DNA damage
DNA damage, due to environmental factors and normal metabolic processes inside the cell, occurs at a rate of 1,000 to 1,000,000 molecular lesions per cell per day.[1] While this constitutes only 0.000165% of the human genome's approximately 6 billion bases (3 billion base pairs), unrepaired lesions in critical genes (such as tumor suppressor genes) can impede a cell's ability to carry out its function and appreciably increase the likelihood of tumor formation.
The vast majority of DNA damage affects the primary structure of the double helix; that is, the bases themselves are chemically modified. These modifications can in turn disrupt the molecules' regular helical structure by introducing non-native chemical bonds or bulky adducts that do not fit in the standard double helix. Unlike proteins and RNA, DNA usually lacks secondary structure and therefore damage or disturbance does not occur at that level. DNA is, however, supercoiled and wound around "packaging" proteins called histones, and both superstructures are vulnerable to the effects of DNA damage.
Sources of damage
DNA damage can be subdivided into two main types:
- endogenous damage such as attack by reactive oxygen species produced from normal metabolic byproducts (spontaneous mutation), especially the process of oxidative deamination;
- exogenous damage caused by external agents such as
- ultraviolet [UV 200-300nm] radiation from the
sun
other radiation frequencies, including x-rays and gamma rays
hydrolysis or thermal disruption
certain plant toxins
human-made mutagenic chemicals, especially aromatic compounds that act as DNA intercalating agents
cancer chemotherapy and radiotherapy
- ultraviolet [UV 200-300nm] radiation from the
sun
The replication of damaged DNA before cell division can lead to the incorporation of wrong bases opposite damaged ones. Daughter cells that inherit these wrong bases carry mutations from which the original DNA sequence is unrecoverable (except in the rare case of a back mutation, for example, through gene conversion).
Types of damage
There are four main types of damage to DNA due to endogenous cellular processes:
- oxidation of bases [e.g. 8-oxo-7,8-dihydroguanine
(8-oxoG)] and generation of DNA strand interruptions
from reactive oxygen species,
alkylation of bases (usually methylation), such as formation of 7-methylguanine
hydrolysis of bases, such as deamination, depurination and depyrimidination.
mismatch of bases, due to DNA replication in which the wrong DNA base is stitched into place in a newly forming DNA strand.
Damage caused by exogenous agents comes in many forms. Some examples are:
- UV light causes crosslinking between adjacent
cytosine and thymine bases creating pyrimidine dimers
Ionizing radiation such as that created by radioactive decay or in cosmic rays causes breaks in DNA strands
Industrial chemicals such as vinyl chloride and hydrogen peroxide, and environmental chemicals such as polycyclic hydrocarbons found in smoke, soot and tar create a huge diversity of DNA adducts- ethenobases, oxidized bases and alkylated phosphotriesters, just to name a few.
Nuclear versus mitochondrial DNA damage
In human, and eukaryotic cells in general, DNA is found in two cellular locations - inside the nucleus and inside the mitochondria. Nuclear DNA (nDNA) exists as chromatin during non-replicative stages of the cell cycle and is condensed into aggregate structures known as chromosomes during cell division. In either state the DNA is highly compacted and wound up around bead-like proteins called histones. Whenever a cell needs to express the genetic information encoded in its nDNA the required chromosomal region is unravelled, genes located therein are expressed, and then the region is condensed back to its resting conformation. Mitochondrial DNA (mtDNA) is located inside mitochondria organelles, exists in multiple copies, and is also tightly associated with a number of proteins to form a complex known as the nucleoid. Inside mitochondria, reactive oxygen species (ROS), or free radicals, byproducts of the constant production of adenosine triphosphate (ATP) via oxidative phosphorylation, create a highly oxidative environment that is known to damage mtDNA. A critical enzyme in counteracting the toxicity of these species is superoxide dismutase, which is present in both the mitochondria and cytoplasm of eukaryotic cells.
Senescence and apoptosis
Senescence, an irreversible state in which the cell no longer divides (mitosis), is a protective response to the shortening of the chromosome ends (telomeres). The telomeres are long regions of repetitive noncoding DNA that cap chromosomes and undergo partial degradation each time a cell undergoes division (see Hayflick limit).[3] In contrast, quiescence is a reversible state of cellular dormancy that is unrelated to genome damage (see cell cycle). Senescence in cells may serve as a functional alternative to apoptosis in cases where the physical presence of a cell for spatial reasons is required by the organism,[4] which serves as a "last resort" mechanism to prevent a cell with damaged DNA from replicating inappropriately in the absence of pro-growth cellular signaling. Unregulated cell division can lead to the formation of a tumor (see cancer), which is potentially lethal to an organism. Therefore the induction of senescence and apoptosis is considered to be part of a strategy of protection against cancer.
DNA repair mechanisms
Cells cannot function if DNA damage corrupts the integrity and accessibility of essential information in the genome (but cells remain superficially functional when so-called "non-essential" genes are missing or damaged). Depending on the type of damage inflicted on the DNA's double helical structure, a variety of repair strategies have evolved to restore lost information. If possible, cells use the unmodified complementary strand of the DNA or the sister chromatid as a template to losslessly recover the original information. Without access to a template, cells use an error-prone recovery mechanism known as translesion synthesis as a last resort.
Damage to DNA alters the spatial configuration of the helix and such alterations can be detected by the cell. Once damage is localized, specific DNA repair molecules are summoned to, and bind at or near the site of damage, inducing other molecules to bind and form a complex that enables the actual repair to take place. The types of molecules involved and the mechanism of repair that is mobilized depend on the type of damage that has occurred and the phase of the cell cycle that the cell is in.
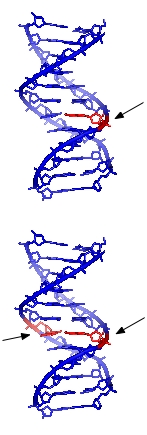
Direct reversal
Cells are known to eliminate three types of damage to their DNA by chemically reversing it. These mechanisms do not require a template, since the types of damage they counteract can only occur in one of the four bases. Such direct reversal mechanisms are specific to the type of damage incurred. The formation of thymine dimers (a common type of cyclobutyl dimer) upon irradiation with UV light results in an abnormal covalent bond between adjacent thymidine bases. The photoreactivation process in bacteria directly reverses this damage by the action of the enzyme photolyase, which uses energy absorbed from UV light to promote catalysis. Another type of damage, methylation of guanine bases, is directly reversed by the protein methyl guanine methyl transferase (MGMT). This is an expensive process because each MGMT molecule can only be used once; that is, the reaction is stoichiometric rather than catalytic.[5] A generalized response to methylating agents in bacteria is known as the adaptive response and confers a level of resistance to alkylating agents upon sustained exposure.[6]The third type of DNA damage reversed by cells is certain methylation of the bases cytosine and adenine.
Single strand damage
When only one of the two strands of a double helix has a defect, the other strand can be used as a template to guide the correction of the damaged strand. In order to repair damage to one of the two paired molecules of DNA, there exist a number of excision repair mechanisms that remove the damaged nucleotide and replace it with an undamaged nucleotide complementary to that found in the undamaged DNA strand.[5]
- Base excision repair (BER), which repairs damage due
to a single nucleotide caused by oxidation, alkylation,
hydrolysis, or deamination;
Nucleotide excision repair (NER), which repairs damage affecting longer strands of 2-30 bases. This process recognizes bulky, helix-distorting changes such as thymine dimers as well as single-strand breaks (repaired with enzymes such UvrABC endonuclease). A specialized form of NER known as Transcription-Coupled Repair (TCR) deploys high-priority NER repair enzymes to genes that are being actively transcribed;
Mismatch repair (MMR), which corrects errors of DNA replication and recombination that result in mispaired nucleotides following DNA replication.
Double strand breaks
A type of DNA damage particularly hazardous to dividing cells is a break to both strands in the double-helix. Two mechanisms exist to repair this damage. They are generally known as non-homologous end-joining (NHEJ) and recombinational repair (also known as template-assisted repair or homologous recombination repair).[5]
The NHEJ pathway operates when the cell has not yet replicated the region of DNA on which the lesion has occurred. The process directly joins the two ends of the broken DNA strands without a template, losing sequence information in the process. Thus this repair mechanism is necessarily mutagenic. However, if the cell is not dividing and has not replicated its DNA, the NHEJ pathway is the cell's only option. NHEJ relies on chance pairings, or microhomologies, between the single-stranded tails of the two DNA fragments to be joined. There are multiple independent "failsafe" pathways for NHEJ in higher eukaryotes.[7]
Recombinational repair requires the presence of an identical or nearly identical sequence to be used as a template for repair of the break. The enzymatic machinery responsible for this repair process is nearly identical to the machinery responsible for chromosomal crossover during meiosis. This pathway allows a damaged chromosome to be repaired using the newly created sister chromatid as a template, i.e. an identical copy that is also linked to the damaged region via the centromere. Double-stranded breaks repaired by this mechanism are usually caused by the replication machinery attempting to synthesize across a single-strand break or unrepaired lesion, both of which result in collapse of the replication fork.
It should be noted that topoisomerases sometimes introduce both single and double strand breaks in the course of changing the DNA's state of supercoiling, which is especially common in regions near an open replication fork. Such breaks are not considered DNA damage because they serve a biochemical purpose and are immediately repaired by the enzymes that created them.
A team of French researchers bombarded Deinococcus radiodurans to study the mechanism of double-strand break DNA repair in that organism. At least two copies of the genome, with random DNA breaks, can form DNA fragments through annealing. Partially overlapping fragments are then used for synthesis of homologous regions through a moving D-loop that can continue extension until they find complementary partner strands. In the final step there is crossover by means of RecA-dependent homologous recombination[8].
Translesion synthesis
Translesion synthesis is an error-prone (almost error-guaranteeing) last-resort method of repairing a DNA lesion that has not been repaired by any other mechanism. The DNA replication machinery cannot continue replicating past a site of DNA damage, so the advancing replication fork will stall on encountering a damaged base. The translesion synthesis pathway is mediated by specific DNA polymerases that insert extra bases at the site of damage and thus allow replication to bypass the damaged base to continue with chromosome duplication. From the cell's perspective, it is "better" to introduce mutations around a single site than to continue the cell cycle with an incompletely replicated chromosome. The bases inserted by the translesion synthesis machinery are template-independent, but not arbitrary; for example, one human polymerase inserts adenine bases when synthesizing past a thymine dimer.[5]
DNA repair and aging
Poor DNA repair induces pathology
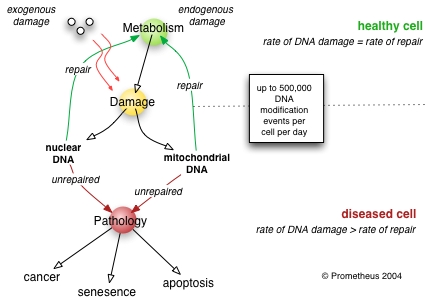
Experimental animals with genetic deficiencies in DNA repair often show decreased lifespan and increased cancer incidence. For example, mice deficient in the dominant NHEJ pathway and in telomere maintenance mechanisms get lymphoma and infections more often, and consequently have shorter lifespans than wild-type mice.[9] Similarly, mice deficient in a key repair and transcription protein that unwinds DNA helices have premature onset of aging-related diseases and consequent shortening of lifespan.[10] However, not every DNA repair deficiency creates exactly the predicted effects; mice deficient in the NER pathway exhibited shortened lifespan without correspondingly higher rates of mutation.[11]
If the rate of DNA damage exceeds the capacity of the cell to repair it, the accumulation of errors can overwhelm the cell and result in early senescence, apoptosis or cancer. Inherited diseases associated with faulty DNA repair functioning result in premature aging, increased sensitivity to carcinogens, and correspondingly increased cancer risk (see below). On the other hand, organisms with enhanced DNA repair systems, such as Deinococcus radiodurans, the most radiation-resistant known organism, exhibit remarkable resistance to the double strand break-inducing effects of radioactivity, likely due to enhanced efficiency of DNA repair and especially NHEJ.[12] It is noteworthy that some workers suguest that if a DNA damage event occurs during the self repair process then the combination of the two events will exert an effect greater than the sum of the individual events (if they occured with a long time delay between them), this is the basis of the second event theory favoured by C. Busby (The Low Level Radiation Campaign).
Longevity and caloric restriction
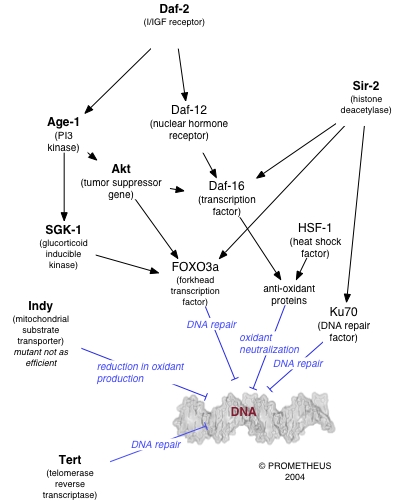
A number of individual genes have been identified as influencing variations in lifespan within a population of organisms. The effects of these genes is strongly dependent on the environment, particularly on the organism's diet. Caloric restriction reproducibly results in extended lifespan in a variety of organisms, likely via nutrient sensing pathways and decreased metabolic rate. The molecular mechanisms by which such restriction results in lengthened lifespan are as yet unclear (see [13] for some discussion); however, the behavior of many genes known to be involved in DNA repair is altered under conditions of caloric restriction.
For example, increasing the gene dosage of the gene SIR-2, which regulates DNA packaging in the nematode worm Caenorhabditis elegans, can significantly extend lifespan.[14] The mammalian homolog of SIR-2 is known to induce downstream DNA repair factors involved in NHEJ, an activity that is especially promoted under conditions of caloric restriction.[15] Caloric restriction has been closely linked to the rate of base excision repair in the nuclear DNA of rodents,[16] although similar effects have not been observed in mitochondrial DNA.[17]
Interestingly, the C. elegans gene AGE-1, an upstream effector of DNA repair pathways, confers dramatically extended lifespan under free-feeding conditions but leads to a decrease in reproductive fitness under conditions of caloric restriction.[18] This observation supports the pleiotropy theory of the biological origins of aging, which suggests that genes conferring a large survival advantage early in life will be selected for even if they carry a corresponding disadvantage late in life.
Medicine and DNA repair modulation
Hereditary DNA repair disorders
Defects in the NER mechanism are responsible for several genetic disorders, including:
- xeroderma pigmentosum: hypersensitivity to
sunlight/UV, resulting in increased skin cancer
incidence and premature aging
Cockayne syndrome: hypersensitivity to UV and chemical agents
trichothiodystrophy: sensitive skin, brittle hair and nails
Mental retardation often accompanies the latter two disorders, suggesting increased vulnerability of developmental neurons.
Other DNA repair disorders include:
- Werner's syndrome: premature aging and retarded
growth
Bloom's syndrome: sunlight hypersensitivity, high incidence of malignancies (especially leukemias).
ataxia telangiectasia: sensitivity to ionizing radiation and some chemical agents
All of the above diseases are often called "segmental progerias" ("accelerated aging diseases") because their victims appear elderly and suffer from aging-related diseases at an abnormally young age.
Other diseases associated with reduced DNA repair function include Fanconi's anemia, hereditary breast cancer and hereditary colon cancer.
DNA repair and cancer
Inherited mutations that affect DNA repair genes are strongly associated with high cancer risks in humans. Hereditary nonpolyposis colorectal cancer (HNPCC) is strongly associated with specific mutations in the DNA mismatch repair pathway. BRCA1 and BRCA2, two famous mutations conferring a hugely increased risk of breast cancer on carriers, are both associated with a large number of DNA repair pathways, especially NHEJ and homologous recombination.
Cancer therapy procedures such as chemotherapy and radiotherapy work by overwhelming the capacity of the cell to repair DNA damage, resulting in cell death. Cells that are most rapidly dividing - most typically cancer cells - are preferentially affected. The side effect is that other non-cancerous but rapidly dividing cells such as stem cells in the bone marrow are also affected. Modern cancer treatments attempt to localize the DNA damage to cells and tissues only associated with cancer, either by physical means (concentrating the therapeutic agent in the region of the tumor) or by biochemical means (exploiting a feature unique to cancer cells in the body).
DNA repair and evolution
An Ancient and Conserved Mechanism
The basic processes of DNA repair are highly conserved among both prokaryotes and eukaryotes and even among bacteriophage (viruses that infect bacteria); however, more complex organisms with more complex genomes have correspondingly more complex repair mechanisms.[19] The ability of a large number of protein structural motifs to catalyze relevant chemical reactions has played a significant role in the elaboration of repair mechanisms during evolution. For an extremely detailed review of hypotheses relating to the evolution of DNA repair, see [20].
The fossil record indicates that single celled life began to proliferate on the planet at some point during the Precambrian period, although exactly when recognizably modern life first emerged is unclear. Nucleic acids became the sole and universal means of encoding genetic information, requiring DNA repair mechanisms that in their basic form have been inherited by all extant life forms from their common ancestor. The emergence of Earth's oxygen-rich atmosphere (known as the "oxygen catastrophe") due to photosynthetic organisms, as well as the presence of potentially damaging free radicals in the cell due to oxidative phosphorylation, necessitated the evolution of DNA repair mechanisms that act specifically to counter the types of damage induced by oxidative stress.
Evolutionary rate as a function of DNA repair rate
When DNA damage is not repaired properly, or is repaired by an error-prone mechanism, mutations are introduced into the genomes of the cell's progeny. When this occurs in a germ line cell that will eventually produce a gamete, the mutation is passed on to the affected organism's offspring. The rate of evolution in a particular species (or, more narrowly, in a particular gene) is a function of the rate of mutation and thus of the accuracy and the rate of the DNA repair pathway and factors that can influence it.[21]
See also
[edit] References
- ^ a b Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J. (2004). Molecular Biology of the Cell, p963. WH Freeman: New York, NY. 5th ed.
- ^ Browner WS, Kahn AJ, Ziv E, Reiner AP, Oshima J, Cawthon RM, Hsueh WC, Cummings SR. (2004). The genetics of human longevity. Am J Med 117(11):851-60.
- ^ Braig M, Schmitt CA. (2006) . Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res 66: 2881-2884.
- ^ Lynch MD. (2006). How does cellular senescence prevent cancer? DNA Cell Biol 25(2):69-78.
- ^ a b c d Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. (2004). Molecular Biology of the Gene, ch. 9 and 10. Peason Benjamin Cummings; CSHL Press. 5th ed.
- ^ Volkert MR. (1988). Adaptive response of Escherichia coli to alkylation damage. Environ Mol Mutagen 11(2):241-55.
- ^ Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. (2003). Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 31(18):5377-88.
- ^ Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006). "Reassembly of shattered chromosomes in Deinococcus radiodurans". NATURE 443 (7111): 569-573. PMID 17006450.
- ^ Espejel S, Martin M, Klatt P, Martin-Caballero J, Flores JM, Blasco MA. (2004). Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep 5(5):503-9.
- ^ de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. (2002). Premature aging in mice deficient in DNA repair and transcription. Science 296(5571):1276-9.
- ^ Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, van der Horst G, Hoeijmakers JH, van Steeg H, Vijg J. (2006). Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res 596(1-2):22-35.
- ^ Kobayashi Y, Narumi I, Satoh K, Funayama T, Kikuchi M, Kitayama S, Watanabe H. (2004). Radiation response mechanisms of the extremely radioresistant bacterium Deinococcus radiodurans.Biol Sci Space 18(3):134-5.
- ^ Spindler SR. (2005). Rapid and reversible induction of the longevity, anticancer and genomic effects of caloric restriction. Mech Ageing Dev 126(9):960-6.
- ^ Tissenbaum HA, Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410(6825):227-30.
- ^ Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305(5682):390-2.
- ^ Cabelof DC, Yanamadala S, Raffoul JJ, Guo Z, Soofi A, Heydari AR. (2003). Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amst.) 2(3):295-307.
- ^ Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. (2004). Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J 18(3):595-7.
- ^ Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. (2000). Evolution of lifespan in C. elegans. Nature 405(6784):296-7.
- ^ Cromie GA, Connelly JC, Leach DR (2001). Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 8(6):1163-74.
- ^ O'Brien PJ. (2006). Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev 106(2):720-52.
- ^ Maresca B, Schwartz JH (2006). Sudden origins: a general mechanism of evolution based on stress protein concentration and rapid environmental change. Anat Rec B New Anat. Jan;289(1):38-46




 216.73.216.133
216.73.216.133 User Stats:
User Stats:
 Today: 0
Today: 0 Yesterday: 0
Yesterday: 0 This Month: 0
This Month: 0 This Year: 0
This Year: 0 Total Users: 117
Total Users: 117 New Members:
New Members:
 216.73.xxx.xxx
216.73.xxx.xxx
 Server Time:
Server Time: